Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Mass Spectrometry Analysis of Novel Stem Cell-Derived Peptides
*Corresponding author: Jonathan RT Lakey, Professor Emeritus University of California Irvine, USA.
Received: April 29, 2022; Published: May 20, 2022
DOI: 10.34297/AJBSR.2022.16.002215
Abstract
Short peptides have been demonstrated to play an important role in modulating transcription, the transmission of biological information, and in restoring the genetic alterations that occur with aging. This paper aims to describe a method of identifying a population of peptides derived from a peptide cocktail formulation. Samples of Nano Organo Peptides (NOP) and Mito Organelles (MO) Peptides derived from both the Central Nervous System (CNS) and the Lungs (LBS) of Specific Pathogen Free (SPF) mammalian rabbits sourced from Charles River Labs were analyzed by mass spectrometry and chromatograms were generated for further examination. The experimentally derived peaks were searched against a database of known proteins to identify the peptides present in each sample. The following report outlines the experimental methods and the results from performing mass spectrometry on various peptides from the company European Wellness (EW).
Keywords: Peptides; Nano Organo Peptides (NOP); Mito Organelles (MO) Peptides; Mass Spectrometry
Introduction
Peptides are linear polymers formed by a series of amino acid residues that are linked together through peptide bonds [1]. Whereas proteins typically contain between 50 and 2000 amino acid residues and have a mean molecular weight between 5.5 and 22kDa, peptides have fewer than 50 residues and a reduced weight compared to that of proteins [1]. Among cells, a unique set of proteins and peptides are produced that have different functions in regulating biological homeostasis [2]. Short peptides have been demonstrated to play an important role in modulating transcription, transmission of biological information, and in restoring the genetic alterations that occur with age [3]. These peptides are signaling molecules that act as regulatory factors through their interaction with DNA and histone proteins. Moreover, the physiological process of aging is highly influenced by the peptidergic regulation of homeostasis and is related to the aging of cells, tissues, and organs.
Peptide therapy aims to renew the strength of the signals received by cells to either induce peptide production or trigger normal signaling processes, thereby rejuvenating and revitalizing tissues as well as the organism as a whole [3-6].
Although the content of peptides is similar between cells, the function and morphology of each cell defines the content of its biologically active substances and its unique ultrastructure’s. Moreover, certain biologically active substances are predominantly synthesized or accumulated in specific tissues. Since the signaling activity and function of peptides is largely based on the cell type, peptide therapy utilizes organ-specific extracts to target diseased or aging tissue. Due to the short lengths of peptides and their low molecular weights, biosynthesis and extraction processes permit them to be mass produced and distributed for use in therapeutic treatments [3]. Through years of research and extensive global practice, MF-Plus has manufactured two products - Nano Organo Peptides (NOPs) and Mito Organelles (MO) Peptides - that are intended for use in both animals and humans as a revitalization therapy [7].
Nano Organo Peptides (NOPs) are 3nm in size and have a molecular weight of less than 10kDA [3]. NOPs are procured from mammalian stem cells and are processed through a proprietary parallel-extraction process that includes multiple ultrafiltrate steps through specialized Millipore’s to obtain the cellular material within the cell, known as the molecular-level ultrafiltrates. Owing to the extraction process, these ultrafiltrates are specific to the cell type that they are derived from. NOP contents are extracted from organ specific cells with an initially high molecular mass and subsequently separated through various ultrafiltration steps through micro-Millipore filters. This selective filtration process only allows substances with a molecular mass of less than 10kDa to pass, thereby ensuring peptide specificity. Moreover, the small molecular weights and high solubility of NOPs permits their delivery via both sublingual and injectable routes (either subcutaneous or intramuscular) [3]. NOPs have been investigated and utilized for a variety of applications including cosmetics [8] and regenerative organ repair [9].
Mito Organo (MO) peptides are biologically extracted mixtures of cellular peptides that have predominantly mitochondria-specific functions [10]. Although cells of different organ systems have similar functions, variations in cellular functions between organs creates the differential expression of peptides, which can be utilized for therapeutic purposes. As part of the aging process, the volume and strength of signals to the mitochondria declines, causing signals to be sent back to the nucleus to arrest cell proliferation and initiate apoptosis and cell death. MO peptides are organ-specific extracts that are aimed at revitalizing and rejuvenating mitochondrial activity, thereby regenerating cells and organisms. Unlike NOPs, MOs are larger in size and have predominantly mitochondriaspecific functions that allows for a more pronounced revitalization of mitochondrial function [11,12].
Despite the numerous studies highlighting the therapeutic effectiveness of NOP and MO peptides and the established procedures documented on obtaining them, little is known of the exact makeup of these formulations. Mass spectrometry has been shown to identify and quantify analytes in complex solution [13] and therefore is thought to be able to identify the population of peptides derived from peptide cocktail formulations. The mass spectrometer produces a readout of peaks plotted in relative abundance against the mass-to-charge ratios. By searching the experimentally derived peaks against a database of known proteins, it is possible to identify the peptides. The following report outlines the experimental methods and the results from performing mass spectrometry on various EW peptides.
Materials and Methods
Eight samples of EW peptides were suspended in saline solution, with two equivalent batches of four formulations labeled: LBS MO201901, CNS MO201901, CNS NOP201901, and LBS NOP201901. Formulations with the characterization LBS were extracted from lung samples and formulations with the characterization CNS were extracted from the central nervous system. All samples were kept on ice throughout the duration of the experiment and were handled using good laboratory practices./p>
Sample Preparation
Samples were refrigerated in sterile bottles until used. The Thermo Fisher Scientific BCA Protein Assay protocol was utilized to determine the protein concentrations of the unknown peptide solutions. Triplicate sample readings were obtained for the four unknown peptide solutions using the Tecan Infinite F200 microplate reader at a wavelength of 570nm.
Preliminary Mass Spectrometry Data Preparation
Peptides were withdrawn from the sterile bottles at a volume of 30μg/mL and pipetted into auto column tubes. Each sample loaded for mass spectrometry (Waters Xevo G2-XS QTof) was diluted with Deionized (DI) water to keep the concentrations of peptide below the overloading dose for the mass spectrometer. The dilution factor was based on the protein concentration determined from the BCA assay. Once samples were loaded in the columns, they were gently vortexed for 1-3 seconds to ensure proper mixing. A total of 26 columns, made up of three replicates per sample and two auto column tubes of 0.9% saline controls, were labeled appropriately loaded onto plates in the mass spectrometer. Sample name, test type, inlet file type, column position on plate, and injection volume of sample were all entered the Mass Lynx software. A blank was used between runs to clear the lines for the next sample as well as to serve as a check for possible contaminants. Once all samples were loaded and sample information was entered into Mass Lynx, the mass spectrometry automation program was started. Chromatograms were generated and stored for analysis using the Mass Lynx software. The sample LBS MO was selected for MADLITOF analysis due to its relatively higher protein concentration and more prominent chromatogram peaks.
Sample Preparation of LBS MO Sample
Peptides from the LBS MO samples were withdrawn at a volume of 30μg/mL and pipetted into auto column tubes in an agarose gel. Sample mixtures were separated by molecular weight with SDSPAGE and Coomassie™ blue staining was used to visualize proteins. An individual protein band was cut from the gel and placed into a low-binding, siliconized microcentrifuge tube. Proteins were then de-stained in the tube with 100μl of a 1:1 methanol and water solution and vortexed. The gel piece was further washed by removing the de-stained solution in the tube and adding 400μl of water. Tubes were shaken for 15mins at room temperature, and the de-staining step was continued until the gel became colorless (minimum of 3 repeats). 400μl of 100% acetonitrile was added to dehydrate the gel for 10mins and dried by vacuum centrifugation after removal of the supernatant. Disulfide bonds were removed with the addition of 100μl of 10mM Dithiothreitol (DTT) to the gel and then the gel was incubated for 45 minutes at 55°C. The solution was removed and 100μl of 55mM iodoacetamide was added to the gel and it was then incubated for an additional 30 minutes at room temperature in low light conditions, allowing trypsin to access cleavage sites. The solution was then added to 400μl of the gel and a wash solution (50% per volume acetonitrile, 25mM ammonium bicarbonate) was added. The gel was then incubated at room temperature and vortexed for 15 minutes three times. The gel was subsequently dehydrated with 400μl of 100% acetonitrile for 10 mins and dried in vacuum centrifuge after removal of the supernatant.
Enzyme Digestion
A protease trypsin solution with a pH 8, diluted 1:1000 with 25mM ammonium bicarbonate, was prepared and diluted to a final concentration of 10-20μg/ml. Trypsin was added to the gel, and it was incubated on ice for 1 hour. The solution was removed and replaced with 25mM ammonium bicarbonate to cover the gel. The gel was then incubated at 37°C overnight.
Peptide Extraction
The supernatant containing the peptides was transferred to a new microcentrifuge tube. A gel extraction solution of 50% per volume acetonitrile, 1% Trifluoroacetic Acid (TFA) and 49% water was added, which was followed by incubation at room temperature and vortexed for 20 minutes. This solution was combined with the supernatant of the peptides in the new microcentrifuge tube.
Analysis
10μl of a 1:1 solution of 0.1% TFA and 100% acetonitrile was added to 10μl of each sample. 1μl of DMP was added onto the MALDI plate. 1μl of sample solution was then applied with 1:1 0.1% TFA and 100% acetonitrile onto the same spot as the DMP and the spot was allowed to dry. The samples were run on MALDI in triplicate and the peaks were compared against a database to identify the key peptides and amino acids found in the samples.
Data Analysis
Data was analyzed using the chromatogram tool in Mass Lynx software. Replicates for each sample were analyzed together and chromatograms for each were generated. Batches 1 and 2 were matched to compare consistency across products. The mass of the most prominent three peaks was deconvoluted and the key peptides and amino acid components were determined against open-source databases.
Results
The purpose of this study was to identify the composition of the unknown peptide solutions in their cellular-derived formulations. The first step involved determining the concentration of peptides (ug/mL) in each solution. To do so, a BCA protein assay was performed on both batches of the four sample groups. As illustrated in Figure 1, all samples were found to have a protein concentration above 150ug/mL and with relatively low variability between batches. Although no statistically significant differences were found within batches (p=ns), protein concentration between samples varied significantly and could be accounted for due to the heterogeneous nature of the cellularly-derived solutions.
The samples underwent mass spectrometry analysis using the Waters Xevo G2-XS QTof spectrometer, a form of LC-MS/MS based peptide sequencing. There was little variability shown between batches, however there was more variability shown between samples (Figure 2). However, the NOP peptide samples from both CNS (Figure 2A) and LBS (Figure 2B) showed near identical chromatograms.
Based on our preliminary mass spectrometry data, the sample LBS MO was selected for further analysis due to its higher protein concentration and the presence of prominent peaks pointing towards the presence of large peptide fragments. Figure 3 illustrates the chromatograms of batches 1 and 2 of LBS MO obtained from MALDI-TOF.

Figure 1: Average protein concentration (ug/mL) of batch 1 and 2 for LBS MO, CNS MO, CNS NOP, and LBS NOP. The black bars represent standard error. No statistically significant differences were found between batches for any samples (p=ns) although statistically significant differences were found between groups (p<0.05).

Figure 2: Chromatogram variability in both batches of samples: MO CNS (A), MO LBS (B), NOP CNS (C), NOP LSBS (D). The green, blue, and red chromatograms associated with both Batches 1 and 2 illustrate different runs of the sample.

Figure 3: Chromatograms obtained from batch 1 (A) and batch 2 (B) of LBS MO. The five red arrows pointing upwards on the x-axis of each chromatogram identify peaks that correspond to primary peptide components of the solution. The green, blue, and red chromatograms associated with both batch 1 and batch 2 illustrate different runs of the sample.
The deconvoluted masses of batch 1 (Figure 4) and batch 2 (Figure 5) of LBS MO also show similarity in major protein components. These masses refer to the red arrows in Figure 3 which show that the higher the peak, the higher concentration of protein is present. However, there was some variation in the smaller protein components between samples 1 and 2. The deconvoluted masses in batch 1 represent five major proteins with sizes of 14,969 Da, 15,300 Da, 8,449 Da, 8,294 Da and 4,618 Da respectively. Batch 2 had six major protein masses of 14,969 Da, 15,301 Da, 8,294 Da, 8,449 Da, 5,436 Da, and 6,214 Da respectively.
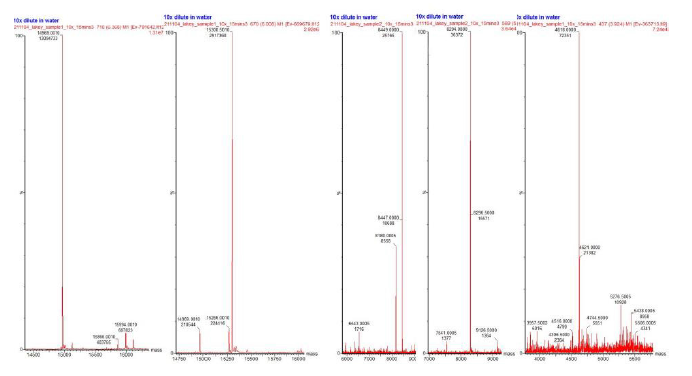
Figure 4: Deconvoluted mass spectrometry data obtained from LBS MO Batch 1. Five peptide fragments are ordered from left to right in terms of abundance within the sample. The relative sizes of peptides from left to right are 14,969 Da, 15,300 Da, 8,449 Da, 8,294 Da and 4,618 Da respectively..

Figure 5: Deconvoluted mass spectrometry data obtained from LBS MO Batch 2. Six peaks representing six major peptide fragments are ordered from left to right in terms of abundance within the sample. The relative sizes of peptides from left to right are 14,969 Da, 15,301 Da, 8,294 Da, 8,449 Da, 5,436 Da, and 6,214 Da respectively.
Discussion and Conclusion
modulation of transcription and transmission of biological information and have been found to decline in production during the natural process of aging [2]. Peptide therapy aims to either induce peptide production or reinstate normal signaling patterns by renewing the strength of the signals received by cells. Due to the differences in peptides produced between different tissue types, peptide therapy utilizes organ-specific extracts to target aging or diseased tissue with the goal of revitalizing normal peptidergic signaling in these regions and improving overall health and wellbeing. Through years of testing and development, European Wellness (EW) and MF-Plus has manufactured two products– Nano Organo Peptides (NOPs) and Mito Organelles (MO) peptides– that are intended for use in peptide therapy in both humans and animals [7]. Despite numerous studies demonstrating the potential of NOPs and MOs in therapeutic applications such as cosmetics [8] and regenerative organ repair [9], little research has been done to investigate and identify the key peptides in these solutions. Mass Spectrometry (MS) is a chemical analysis technique that enables the direct identification of molecules based on their mass-tocharge ratios and fragmentation patterns [14]. By comparing experimental MS data with that of well-established open-source databases, a determination can be made of the proposed identity of the molecules, peptides, or proteins found within a solution. Due to the low-cost and rapid application of MS in identifying the components of unknown solutions, our study employed MS as our primary method of identification.
Prior to the commencement of an in-depth analysis to identify key protein components of one of the solutions, a preliminary study was conducted to determine the relative protein concentrations of each sample (Figure 1), the variability between batches (Figure 2), and to identify which solution demonstrated the most peaks of potential interest. Our results indicated that there were no statistically significant differences in protein concentration (ug/ mL) between batches but were statistically significant differences between sample types (p<0.05). These results are in line with our experimental expectations as both the NOP and MO samples are cellularly-derived solutions that contain a heterogenous mixture of molecules and peptides that will vary depending on the tissue that they are recovered from. Between batches, however, relative protein concentrations were expected to remain consistent as they were procured from the same tissue samples and processed in the same manner.
In addition to the consistency between batches found in the protein concentration assay, consistency was also demonstrated between batches during our preliminary MS experiment (Figure 2). Following our preliminary MS experiment and protein concentration assay, LBS MO was chosen to progress and to perform an in-depth analysis due to its relatively high protein concentration and large number of peaks that indicate potential peptide candidates. Additionally, due to the differences in extraction and preparation procedures between the MO and NOP sample, the MO sample was deemed to be a more desirable candidate due to the likelihood of finding peptides of larger size and weight.
Although our preliminary data relied upon LC-MS/MS based peptide sequencing techniques to produce chromatograms and deconvolute our data, we utilized MALDI-TOF MS identification techniques for our in-depth analysis due to several calculated benefits [15]. First, MALDI-TOF utilizes a protein fingerprinting method in which the sample is digested by a proteolytic enzyme such as trypsin and used to generate an MS spectrum that can be searched against a database. Matched hits are ranked according to a scoring method in which the candidate protein that contains more proteolytic peptides has a higher score and generally represents the protein/peptide that is most probable. The desirability of MALDITOF also includes the speed at which each run is performed-often less than one minute to obtain-and the speed at which analysis can be performed against a database. In contrast, data acquired through LC-MS/MS typically requires multiple hours to obtain due to the length of runs-sometimes requiring over thirty minutes for each run-and requires several hours to parse through the data to identify a potential candidate. Unlike MALDI-TOF, LC-MS/MS requires data to be manually sorted and compared against a database. This can be time consuming and won’t necessarily guarantee the most accurate result.
The results of our study using MALDI-TOF to analyze the MO LBS sample indicated that there were five major peptide products of interest in batch 1 and six major peptide products in batch 2 (Figures 4 & 5). Following deconvolution, five key peptides were identified in batch 1 (Figure 4) with masses of 14,969 Da, 15,300 Da, 8,449 Da, 8,294 Da and 4,618 Da. Batch 2 identified four of the same peptides-14,969 Da, 15,301 Da, 8,294 Da, 8,449 Da in size and two peptides of 5,436 Da, and 6,214 Da in size (Figure 5). Slight differences in peptide products between batches is likely due to the heterogeneous nature of cellularly-derived solutions and differences that occurred during the extraction process. Further research must be conducted to confirm or disprove this theory.
Author Contributions
Conceptualization, J.L., M.W., M.C., D.K., D.C.; methodology, A.W. A.G., B.K., M.A.; validation, A.W., A.G., B.K.; formal analysis, A.W., A.G., B.K.; investigation, A.W., A.G., B.K.; data curation, A.W., A.G., B.K.; writing-original draft preparation, A.W., A.G., K.D.; writingreview and editing, A.W., A.G., J.L.; visualization, J.L., M.W., M.C., D.K.; supervision, J.L., M.W., M.C., D.K., D.C., B.K., A.M.; project administration, J.L., M.W., M.C., D.K., D.C., B.K.; funding acquisition, J.L., M.W., M.C., D.K., D.C.. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant to the University of California, Irvine from BioPep.
Institutional Review Board Statement
Not applicable
Data Availability Statement
The data presented in this study are available in the study outlined.
Acknowledgments
The authors wish to acknowledge the support of the Department of Surgery and the Core Mass Spectrometry laboratory of the University of California, Irvine for their support.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Berg JM, Tymoczko JL, Stryer L (2002) Primary Structure: Amino Acids Are Linked by Peptide Bonds to Form Polypeptide Chains. In Biochemistry 5.
- Caputi S, Trubiani O, Sinjari B, Trofimova S, Diomede F, et al. (2019) Effect of short peptides on neuronal differentiation of stem cells. Int J Immunopathol Pharmacol 33: 2058738419828613.
- Khavinson VK, Lin'kova NS, Tarnovskaya SI (2015) Short Peptides Regulate Gene Expression. Bulletin of experimental biology and medicine 162(2): 288-292.
- Klokol D, Nallenthiran L, Chan MKS, Wong MBF, Chernykh V, et al. (2019) Cell therapy as the main stratagem of anti-aging and regenerative medicine. Europ J Pharm Med Res 6(6): 295-299.
- Lee AC, Harris JL, Khanna KK, Hong JH (2019) A comprehensive review on current advances in peptide drug development and design. International journal of molecular sciences 20(10): 2383.
- De La Torre BG, Albericio F (2020) Peptide therapeutics 2.0. Molecules (Basel, Switzerland) 25(10): 2293.
- Klokol D, Chan M, Wong M, Tullina D, Chernykh, V, et al. (2017) Pathogenetically based integrative therapeutic strategies in management of autism spectrum disorders. Journal of Stem Cell Research and Medicine 2(2): 1-5.
- Chan MKS, Wong MBF, Béguin A, Teppone M, Tukhvatullina D, et al. (2016) In Efficacy of the MFIII placena extracts soft gels supplementation: a randomized double-blind placebo-controlled study. Basic Research Journal of Medicine and Clinical Sciences 5(4): 86-95.
- Chan MK, Michelle WB, Klokol D, Pong H, Wolodymyr C (2017) Biohormonal therapyin early management of premature menopause and andropause. International Journal of Current Medical and Pharmaceutical Research 3(1): 1278-1281.
- Lee C, Zeng J, Drew BG, Sallam T, Martin Montalvo A, et al. (2016) The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell metabolism 21(3): 443-454.
- Wu D, Kampmann E, Qian G (2021) Novel insights into the role of mitochondria-derived peptides in myocardial infarction. Frontiers in Physiology 12.
- Krejcova G, Patocka J, Slaninova J (2004) Effect of humanin analogues on experimentally induced impairment of spatial memory in rats. J Pept Sci 10: 636-639.
- Hopfgartner Gr, Bourgogne E (2003) Quantitative high-throughput analysis of drugs in biological matrices by mass spectrometry. Mass Spectrometry Reviews 22(3): 195-214.
- Urban PL (2016) Quantitative mass spectrometry: an overview. Philosophical transactions. Series A, Mathematical, physical, and engineering sciences 374(2079): 20150382.
- Padoan A, Basso D, Zambon CF, Prayer Galetti T, Arrigoni G, et al. (2018) MALDI-TOF peptidomic analysis of serum and post-prostatic massage urine specimens to identify prostate cancer biomarkers. Clin Proteomics 15: 23.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.